5. Animal Tracking Data Analysis¶
Organism tracking is performed through the MCAM GUI resulting in .CSV output files containing tracking data. This page explores concepts relating to loading data from the CSVs, plotting data, and filtering data.
Output Files¶
Output files are saved in a folder with the same name as the input file and this folder is created in the parent directory of the input file.
Raw Tracking Data - tracking_data.csv - CSV file
containing 8 key-points per fish, x, y coordinate and
confidence for each keypoint. Note: This file is likely
too large to be opened in Excel or other spreadsheet
software due to the large number of columns in the
data array. This file can be opened and manipulated
using Python.
Plotted Tracks -
plotted_tracks.png- Visualization of movement over time from blue (earliest, cold) to red (most recent, hot).Distance Traveled Data -
distance_traveled_metrics.csv- Distance traveled and speed calculated for each frame.Aggregate Metrics -
distance_traveled_key_metrics.csv- Distance traveled and speed calculated for each frame.Video Composite -
composite_tracked_video.mp4- MP4 video containing key-point and skeleton labeled fish for all wells of a well plate.
Units¶
Unless otherwise specified, exported units implicitly:
Distance or length: meters
Time: seconds
Velocity or speed: meters/second
Orientation/Angle: degrees
Zebrafish Examples¶
Zebrafish Tracking Workflow¶
1"""
2# %% Sample zebrafish tracking workflow script
3# By Ramona Optics Inc. Copyright 2023-2025
4
5This script provides an example to walk through the steps of the tracking workflow.
6
7"""
8
9from pathlib import Path
10
11from tqdm import tqdm
12
13from owl import mcam_data
14from owl.analysis.models import fetch_model, zebrafish_models_recommended
15from owl.analysis.tracking import infer_dataset
16from owl.analysis.tracking_data_analysis import (
17 compute_fish_length,
18 export_csv,
19 generate_tracking_dataset,
20 make_dataframe,
21)
22from owl.visualize.tracking import plot_tracks_from_dataset
23
24# %% define the project path where outputs are collected
25exported_path = '/MCAM_data/EXPORTED_FOLDER_YOU_WANT_TO_TRACK'
26
27# %% load the exported dataset video file
28video_dataset = mcam_data.load(exported_path)
29
30# %% determine the well plate configuration, i.e. 24, 48 or 96 well plate
31well_plate = (int(video_dataset.wellplate_config_rows) *
32 int(video_dataset.wellplate_config_columns))
33
34# %% create a results folder within the output data path
35tracking_filepath = Path(exported_path)
36results_folder = tracking_filepath / 'results'
37results_folder.mkdir(parents=True, exist_ok=True)
38
39# %% download and point to a pre-trained tracking model
40recommended_model_name = zebrafish_models_recommended[f"{well_plate}_well_plate"]
41model_path = fetch_model(recommended_model_name)
42
43tracking_dataset = infer_dataset(
44 video_dataset,
45 model_path,
46 tqdm=tqdm,
47)
48
49# %% make computations based on raw tracking data and store them in the dataset
50tracking_dataset['fish_length_information'] = compute_fish_length(tracking_dataset)
51
52# %% plot the tracks on representative images, color gradient represents speed
53plot_tracks_from_dataset(
54 tracking_dataset,
55 output_filename=results_folder / 'plotted_tracks.png',
56 apply_circle_mask=True,
57 apply_square_mask=False,
58 speed=True,
59)
60
61# %% save the tracking data separately from the dataset for future analysis
62# the images are already saved so we do not save them again
63tracking_dataset = tracking_dataset.drop_vars('images')
64tracking_dataset.to_netcdf(tracking_filepath / 'tracking_metadata.nc')
65
66# %% prepare tracking dataset for export to csv format including unit conversions
67tracking_dataset = generate_tracking_dataset(tracking_dataset)
68
69# %% prepare raw tracking data for export
70tracking_output_df, header = make_dataframe(
71 tracking_dataset,
72 information_name='tracking_information',
73 row_index='time',
74 column_indices=('well_row_letter',
75 'well_column_number',
76 'tracking_keypoint',
77 'tracking_location'),
78)
79# %% save the raw tracking data
80export_csv(
81 tracking_output_df,
82 results_folder / 'tracking_data.csv',
83 header=header,
84 index=True
85)
Tracking Dataset - Data Filtering¶
1"""
2# %% Sample zebrafish tracking workflow script
3# By Ramona Optics Inc. Copyright 2023-2025
4
5This script provides an example to filter tracking data using
6anomaly detection and denoising.
7
8"""
9
10from pathlib import Path
11
12from owl.analysis.tracking_data_analysis import (
13 compile_derived_metrics,
14 compute_fish_length,
15 export_csv,
16 filter_anomalies,
17 generate_tracking_dataset,
18 get_anomaly_mask_well_radius,
19 load_tracking_dataset,
20 make_dataframe,
21)
22from owl.visualize.tracking import plot_tracks_from_dataset
23
24# %% define the project path where outputs are collected
25exported_path = '/MCAM_data/EXPORTED_FOLDER_YOU_WANT_TO_TRACK'
26
27# %% create a results folder within the output data path
28tracking_filepath = Path(exported_path)
29results_folder = tracking_filepath / 'results'
30results_folder.mkdir(parents=True, exist_ok=True)
31
32# %% load the previously tracked dataset
33tracking_dataset = load_tracking_dataset(exported_path)
34
35# %% fish length must be computed prior to anomaly detection
36tracking_dataset['fish_length_information'] = compute_fish_length(tracking_dataset)
37
38# %% determine the well plate configuration, i.e. 24, 48 or 96 well plate
39well_plate = (int(tracking_dataset.wellplate_config_rows) *
40 int(tracking_dataset.wellplate_config_columns))
41
42# %% detect anomalous data, remove it and interpolate to replace missing data
43anomaly_mask_well_radius = get_anomaly_mask_well_radius(
44 well_plate, apply_circle_mask=True,
45)
46tracking_dataset['tracking_information'][...], filtering_information = filter_anomalies(
47 tracking_dataset, well_radius=anomaly_mask_well_radius
48)
49tracking_dataset['filtering_information'] = filtering_information
50
51# %% plot the tracks on representative images, color gradient represents speed
52plot_tracks_from_dataset(
53 tracking_dataset,
54 output_filename=results_folder / 'plotted_tracks.png',
55 apply_circle_mask=True,
56 apply_square_mask=False,
57 speed=True,
58)
59
60# %% convert the dataset from pixel coordinates to SI units
61tracking_dataset = generate_tracking_dataset(tracking_dataset)
62
63# %% compute distance traveled and speed metrics with denoising
64tracking_dataset, denoising_information = compile_derived_metrics(
65 tracking_dataset,
66 denoise=True,
67)
68tracking_dataset['denoising_information'] = denoising_information
69
70# %% prepare movement metric data for export
71metrics_output_df, header = make_dataframe(
72 tracking_dataset,
73 information_name='movement_metrics',
74 row_index='time',
75 column_indices=('well_row_letter',
76 'well_column_number',
77 'tracking_metrics',),
78)
79
80# %% save the distance traveled data
81export_csv(metrics_output_df,
82 results_folder / 'distance_traveled_metrics.csv',
83 header=header,
84 index=True)
Tracking Dataset - Time Binning¶
1"""
2# %% Sample zebrafish tracking workflow script
3# By Ramona Optics Inc. Copyright 2023-2025
4
5This script is an example to bin tracking data by one second intervals.
6
7"""
8
9from pathlib import Path
10
11from owl.analysis.tracking_data_analysis import (
12 compile_derived_metrics,
13 export_csv,
14 generate_tracking_dataset,
15 load_tracking_dataset,
16 make_dataframe,
17)
18
19# %% define the project path where outputs are collected
20exported_path = '/MCAM_data/EXPORTED_FOLDER_YOU_WANT_TO_TRACK'
21
22# %% time bin given in seconds
23time_bin = 1
24
25# %% load a previously tracked dataset
26tracking_dataset = load_tracking_dataset(exported_path)
27
28# %% ensure a results folder exists within the output data path
29tracking_filepath = Path(exported_path)
30results_folder = tracking_filepath / 'results'
31results_folder.mkdir(parents=True, exist_ok=True)
32
33# %% prepare tracking dataset for export including unit conversions
34tracking_dataset = generate_tracking_dataset(tracking_dataset)
35
36# %% calculate distance traveled and speed. this function should be
37# run after 'generate_tracking_dataset' to ensure SI units
38tracking_dataset = compile_derived_metrics(tracking_dataset)
39
40# %% prepare movement metric data for export
41metrics_output_df, header = make_dataframe(
42 tracking_dataset,
43 information_name='movement_metrics',
44 row_index='time',
45 column_indices=('well_row_letter',
46 'well_column_number',
47 'tracking_metrics',),
48 time_bin=time_bin,
49)
50
51# %% save the distance traveled data
52export_csv(metrics_output_df,
53 results_folder / 'distance_traveled_metrics.csv',
54 header=header,
55 index=True)
Tracking Dataset - Data Manipulation¶
1# %%
2# The tracking_metadata.nc is an xarray dataset containing information
3# about the MCAM Acquisition, as well as the information about the automated
4# tracking of zebrafish keypoints and skeletons.
5import xarray as xr
6
7# %%
8# input the path to the tracking metadata file in question
9# a sample dataset is available for download here:
10# https://drive.google.com/file/d/1sN3gFrqNbS2rNeBnw5CUlA46cOvfSYRn/view?usp=share_link
11tracking_metadata_filepath = '/path/to/tracking_metadata.nc'
12# load the tracking dataset
13tracking_dataset = xr.open_dataset(tracking_metadata_filepath)
14
15# %%
16# the tracking pipeline is based on the x, y coordinates of each keypoint
17# first access the location of the zebrafish "center" keypoint
18# in the fourth frame of the video tracked for well 'B4'
19# note frame indexing begins at frame 0 so the fourth frame is "3" by index
20frame_number = 3
21well_letter = 'B'
22well_number = 4
23keypoint = 'center'
24
25center_x = tracking_dataset.tracking_information.sel({
26 'frame_number': frame_number,
27 'image_x': well_letter,
28 'image_y': well_number,
29 'tracking_keypoint': keypoint,
30 'tracking_location': 'x',
31}).data
32center_y = tracking_dataset.tracking_information.sel({
33 'frame_number': frame_number,
34 'image_x': well_letter,
35 'image_y': well_number,
36 'tracking_keypoint': keypoint,
37 'tracking_location': 'y',
38}).data
39# within the tracking dataset, locations are in units of pixels
40# make sure to cast the stored floating point number to an integer
41# so that x, y coordinates can properly register in the 2D image matrix
42print(f'Center Key Point Location: ({int(center_x)}, {int(center_y)})')
43
44# %%
45# note the shape of the tracking data
46# this 5-dimensional data array includes:
47# (N_frames, camera_sensor_y, camera_sensor_x, tracking_keypoints, (y, x, confidence))
48shape = tracking_dataset.tracking_information.shape
49print(f'Tracking dataset shape: {shape}')
50# select each one of these dimensions for examination by it's index in the shape tuple
51print(f'There are {shape[0]} frames in this dataset.')
52print(f'There are {shape[1]} well plate numbers and thus columns in this dataset.')
53print(f'There are {shape[2]} well plate letters and thus rows in this dataset.')
54print(f'Therefore it is a {shape[1] * shape[2]}-well plate.')
55print(f'There are {shape[3]} tracked keypoints in this dataset.')
56print(f'There are {shape[4]} tracking locations in this dataset.')
57
58# %%
59# assuming the skeleton has been computed and compiled into the tracking dataset
60# we can access the length of one tail segment for well 'B4' for the fourth frame
61# here we access the first segment in the 'segment_names' list which is defined by
62# the 'center' and 'between_center_and_mid' key points
63tail_segment_names = [
64 'center_between_center_and_mid',
65 'between_center_and_mid_mid_tail',
66 'mid_tail_between_mid_and_caudal',
67 'between_mid_and_caudal_caudal_fin',
68]
69center_between_center_and_mid_length = tracking_dataset.skeleton_information.sel({
70 'frame_number': frame_number,
71 'image_x': well_letter,
72 'image_y': well_number,
73 'skeleton_segments': tail_segment_names[0],
74 'skeleton_parameters': 'length',
75}).data
76print(f'Length: {center_between_center_and_mid_length} pixels.')
77
78# %%
79# to convert from units of pixels to meters, first access the width of
80# each pixel stored in the dataset, this value is in units of meters
81pixel_width = tracking_dataset['pixel_width'].data
82print(f'Pixel width: {pixel_width} meters.')
83
84# %%
85# convert the length of the tail segment to meters
86tail_segment_length_in_meters = center_between_center_and_mid_length * pixel_width
87print(f'Length in meters: {tail_segment_length_in_meters}')
88
89# %%
90# on this scale it is likely millimeters make more sense than meters
91# convert to millimeters
92tail_segment_length_in_mm = tail_segment_length_in_meters * 1E3
93print(f'Length in millimeters: {tail_segment_length_in_mm}')
94
95# %%
96# output a list of all skeleton segments by segment name
97skeleton_segments = tracking_dataset.skeleton_information.skeleton_segments.data
98print(f'Skeleton segments: {skeleton_segments}')
99# the 'tail_segment_names' list above can be redefined by slicing
100# this list of segments to just the last four, which are all tail segments
101tail_segment_names = skeleton_segments[4:]
102print(f'Tail skeleton segments: {tail_segment_names}')
103# output a list of all skeleton parameters for each skeleton segment
104skeleton_parameters = tracking_dataset.skeleton_information.skeleton_parameters.data
105print(f'Skeleton parameters: {skeleton_parameters}')
106
107# %%
108# access the length of all four tail segments for the fourth frame and sum them
109# to compute the tail length of the zebrafish
110tail_segment_lengths = tracking_dataset.skeleton_information.sel({
111 'frame_number': frame_number,
112 'image_x': well_letter,
113 'image_y': well_number,
114 'skeleton_segments': tail_segment_names,
115 'skeleton_parameters': 'length',
116}).data
117tail_length = tail_segment_lengths.sum()
118print(f'Tail length: {tail_length} pixels.')
119
120# %%
121# get this tail length for the first ten frames and take the average as
122# the computed tail length
123# convert from pixels to millimeters and round this value to 2 decimal places
124tail_segment_lengths = tracking_dataset.skeleton_information.sel({
125 'frame_number': slice(0, 10),
126 'image_x': well_letter,
127 'image_y': well_number,
128 'skeleton_segments': tail_segment_names,
129 'skeleton_parameters': 'length',
130}).data
131tail_lengths = tail_segment_lengths.sum(axis=1)
132average_tail_length_pixels = tail_lengths.mean()
133average_tail_length_millimeters = average_tail_length_pixels * pixel_width * 1E3
134average_tail_length_millimeters = round(average_tail_length_millimeters, 2)
135print(f'Average tail length computed across '
136 f'10 frames: {average_tail_length_millimeters} millimeters.')
137
138# %%
139# Note: in addition to keypoint tracking data and skeleton information
140# other computed values can be accessed similarly from the xarray dataset
141# examples:
142
143# tail angles for the four tail keypoints for well 'B4' in the fourth frame
144# angles are in units of degrees
145# positive (+) reflects tail deflection to the left of the body axis
146# negative (-) reflects tail deflection to the right of the body axis
147tail_angles = tracking_dataset.tail_information.sel({
148 'frame_number': frame_number,
149 'image_x': well_letter,
150 'image_y': well_number,
151 'tail_parameters': 'angle',
152}).data
153print(f'Tail angles: {tail_angles} degrees.')
Zebrafish Stimulus Analysis Plotting¶
1"""
2%% Sample zebrafish analysis for stimulus visualization
3By Ramona Optics Inc. Copyright 2022-2025
4
5This script provides and example to import previously tracked
6locomotion data, averages together the distance traveled for
7each fish, and plots this metric with consideration for where
8stimuli occurred during the experiment. If no stimuli information
9is present, the average distance traveled will be simply plotted.
10
11An additional consideration outlined here is two levels of filtering
12excluding fish or frames from the analysis that were missed by the
13tracking algorithm. In this example, if more than 5% of the total
14frames were missed during tracking, the fish will be excluded.
15
16"""
17from pathlib import Path
18
19import numpy as np
20import pandas as pd
21from matplotlib import pyplot as plt
22
23from owl import mcam_data
24
25# %% definitions:
26# define the filepath location of the previously output tracking data
27tracking_filepath = 'path/tracking_data'
28
29tracking_filepath = Path(tracking_filepath)
30# define the filepath to the distance traveled metrics output .csv.
31distance_speed_filename = tracking_filepath / 'results/distance_traveled_metrics.csv'
32# define the filepath to the raw tracking data for tracking
33# confidence information.
34tracking_data_filename = tracking_filepath / 'results/tracking_data.csv'
35# define an output path for the plot,
36# this should include a filename with '.png' or other image-type suffix
37plot_output_path = tracking_filepath / 'results/distance_traveled_plot.png'
38
39# %% script below
40# load the previously extracted dataset metadata which exists within the
41# tracking filepath
42metadata = mcam_data.load(tracking_filepath / 'metadata.nc')
43
44# load the previously exported distance traveled data
45distance_speed_df = pd.read_csv(
46 distance_speed_filename,
47 comment='#',
48 header=[0, 1, 2],
49 index_col=0,
50)
51# load the previously exported raw tracking data
52tracking_data_df = pd.read_csv(
53 tracking_data_filename,
54 comment='#',
55 header=[0, 1, 2, 3],
56 index_col=0,
57)
58
59# determine the rows and columns that exist in the dataset
60# ensure that the column numbers are interpreted as strings, not integers
61rows = metadata.image_x.data
62columns = metadata.image_y.data.astype('str')
63
64# %% check if stimuli information exists in the metadata
65if 'stimuli_flash_index' in metadata.dims:
66 stimuli_durations = metadata.stimuli_flash_duration.data
67 stimuli_times = metadata.stimuli_flash_start_time.data
68 stimuli_intensities = metadata.stimuli_flash_lux.data
69 stimuli_colors = metadata.stimuli_flash_color.data
70 stimuli_index = metadata.stimuli_flash_index.data
71 print('flash stimulus data loaded!')
72elif 'stimuli_vibrate_index' in metadata.dims:
73 stimuli_durations = metadata.stimuli_vibrate_duration.data
74 stimuli_times = metadata.stimuli_vibrate_start_time.data
75 stimuli_frequencies = metadata.stimuli_vibrate_frequency.data
76 stimuli_index = metadata.stimuli_vibrate_index.data
77 print('vibration stimulus data loaded!')
78else:
79 print('stimulus information does not exist in this dataset!')
80
81# %% filter out fish that have low confidence in tracking and were
82# likely missed by the tracking algorithm
83confidence_threshold = 0.1
84tracked_keypoint = 'center'
85total_frames = len(tracking_data_df)
86confidence_column_keys = list(pd.MultiIndex.from_product(
87 [rows, columns, [tracked_keypoint], ['likelihood']]
88))
89confident_wells = []
90for column_key in confidence_column_keys:
91 confident_data = tracking_data_df[column_key][tracking_data_df[column_key] >=
92 confidence_threshold]
93 confident_frames = len(confident_data)
94 confident_fraction = confident_frames / total_frames
95 percent_confident = round(confident_fraction * 100, 2)
96 if confident_fraction >= 0.95:
97 confident_wells.append(column_key)
98 else:
99 print(f"Well {column_key[0] + column_key[1]} has been excluded "
100 f"with {percent_confident}% confident frames")
101
102# %% average together the distance traveled for all fish with
103# confident tracking
104distance_traveled_column_keys = []
105for well_key in confident_wells:
106 distance_traveled_column_keys.append(
107 (well_key[0], well_key[1], 'distance_traveled')
108 )
109
110average_distance_traveled = \
111 distance_speed_df[distance_traveled_column_keys].mean(axis=1)
112
113time = np.array(distance_speed_df.index)
114
115# %% plot the results
116fig, ax = plt.subplots()
117# plot distance traveled converting from meters to millimeters
118ax.plot(time, average_distance_traveled * 1E3,
119 color='black', label='distance traveled')
120ax.set_ylabel("Average Distance Traveled (mm/s)")
121ax.set_xlabel("Time (s)")
122ax.set_title("Average Distance Traveled")
123ax.legend()
124ax.grid()
125
126if 'stimuli_flash_index' in metadata.dims or 'stimuli_vibrate_index' in metadata.dims:
127 # for each stimulus draw lines at the beginning and end of the stimulus
128 for i in range(len(stimuli_index)):
129 stimulus_start = stimuli_times[i]
130 stimulus_end = stimuli_times[i] + stimuli_durations[i]
131 ax.axvspan(stimulus_start, stimulus_end,
132 alpha=0.2, color='#0e6c67')
133
134# save the plot
135fig.savefig(plot_output_path, dpi=300)
Zebrafish Movement Analysis Plotting¶
1# %% Sample zebrafish analysis for episode detection
2# By Ramona Optics Inc. Copyright 2022-2025
3import numpy as np
4import pandas as pd
5from matplotlib import pyplot as plt
6
7# You can enter the full name of the file yo uwant to track here.
8tracking_data_filename = 'tracking_data.csv'
9distance_speed_filename = 'distance_traveled_metrics.csv'
10
11
12wellplate_diameter = 6.85E-3
13wellplate_radius = wellplate_diameter / 2
14
15tracking_data_df = pd.read_csv(
16 tracking_data_filename,
17 comment='#',
18 header=[0, 1, 2, 3],
19 index_col=0,
20)
21
22distance_speed_df = pd.read_csv(
23 distance_speed_filename,
24 comment='#',
25 header=[0, 1, 2],
26 index_col=0,
27)
28# %% Extract the time, it matches between the two csv files
29time = np.asarray(tracking_data_df.index)
30# %% Select the information we want to extract
31well_name = "B6"
32keypoint = "center"
33
34well_letter = well_name[0]
35well_number = well_name[1:]
36# %%
37
38well_data = tracking_data_df[well_letter, well_number, keypoint]
39yx = well_data[["y", "x"]]
40
41# %%
42fig, ax = plt.subplots()
43
44# Units of x and y are in meters. for a 96 well plate, we can plot them in mm
45ax.plot(yx["x"] * 1E3, yx["y"] * 1E3, '.-', label="RAW")
46ax.add_patch(
47 plt.Circle((0, 0), radius=wellplate_radius * 1E3,
48 edgecolor='Black', facecolor=None, fill=False))
49ax.axis('equal')
50ax.set_xlabel("x (mm)")
51ax.set_ylabel("y (mm)")
52ax.set_title(f"Center position well {well_name} -- Raw (unprocesssed) tracking data")
53ax.set_ylim([-(int(wellplate_radius * 1E3) + 1), int(wellplate_radius * 1E3) + 1])
54
55
56# %% Filter away points that are missed in the analysis
57likelihood_threshold = 0.1
58likelihood = well_data["likelihood"]
59likelihood_below_threshold = likelihood < likelihood_threshold
60
61yx_filtered = yx.to_numpy().copy()
62yx_filtered[likelihood_below_threshold, ...] = np.nan
63
64
65def interpolate_nan_points(points_vector):
66 # https://stackoverflow.com/questions/6518811/interpolate-nan-values-in-a-numpy-array
67 nans = np.isnan(points_vector)
68
69 def f():
70 return lambda z: z.nonzero()[0]
71
72 points_vector[nans] = np.interp(f()(nans), f()(~nans), points_vector[~nans])
73 return points_vector
74
75
76yx_filtered[:, 0] = interpolate_nan_points(yx_filtered[:, 0])
77yx_filtered[:, 1] = interpolate_nan_points(yx_filtered[:, 1])
78
79fig, ax = plt.subplots()
80
81ax.plot(yx_filtered[:, 1] * 1E3, yx_filtered[:, 0] * 1E3, '-',
82 color='#ff7f0e', label="Filtered")
83ax.add_patch(
84 plt.Circle((0, 0), radius=wellplate_radius * 1E3,
85 edgecolor='Black', facecolor=None, fill=False))
86ax.axis('equal')
87ax.set_xlabel("x (mm)")
88ax.set_ylabel("y (mm)")
89ax.set_title(f"Center position well {well_name} -- Filtered tracking data")
90ax.set_ylim([-(int(wellplate_radius * 1E3) + 1), int(wellplate_radius * 1E3) + 1])
91# %% Plot tracks
92fig, ax = plt.subplots()
93
94# Units of x and y are in meters. for a 96 well plate, we can plot them in mm
95ax.plot(yx["x"] * 1E3, yx["y"] * 1E3, '.-', label="RAW")
96ax.plot(yx_filtered[:, 1] * 1E3, yx_filtered[:, 0] * 1E3, '-',
97 label="Filtered")
98ax.add_patch(
99 plt.Circle((0, 0), radius=wellplate_radius * 1E3,
100 edgecolor='Black', facecolor=None, fill=False))
101ax.axis('equal')
102ax.set_xlabel("x (mm)")
103ax.set_ylabel("y (mm)")
104ax.set_title(f"Center position well {well_name} -- Raw and Filtered data")
105ax.set_ylim([-(int(wellplate_radius * 1E3) + 1), int(wellplate_radius * 1E3) + 1])
106ax.legend()
107# %% Extract peak speed
108speed = distance_speed_df[well_letter, well_number, "speed"]
109max_speed = speed.max()
110max_speed_mm_per_s = max_speed * 1E3
111print(f"Maximum speed for well {well_name} = {max_speed_mm_per_s:.2f} mm/s")
112
113# %% Plot the speed over time.
114
115fig, ax = plt.subplots()
116ax.plot(time, speed * 1E3)
117ax.set_ylabel("Speed (mm/s)")
118ax.set_xlabel("Time (s)")
119ax.set_title(f"Speed for zebrafish in well {well_name}")
120ax.grid()
121# %% Threshold the speed
122
123# units of speed for analysis are m/s
124speed_threshold = 30E-3
125index_above_threshold = speed > speed_threshold
126
127fig, ax = plt.subplots()
128ax.plot(time, speed * 1E3, label="Zebrafish Speed (mm/s)")
129ax.plot([time[0], time[-1]], [speed_threshold * 1E3, speed_threshold * 1E3],
130 '--r', label=f"Threshold: {speed_threshold * 1E3:.1f} mm/s")
131ax.plot(time[index_above_threshold], speed[index_above_threshold] * 1E3,
132 '.', label="Speed Above Threshold")
133ax.set_ylabel("Speed (mm/s)")
134ax.set_xlabel("Time (s)")
135ax.set_title(f"Speed for zebrafish in well {well_name}")
136ax.legend()
137ax.grid()
138
139# %% cleanup the data
140# Require at least 5 time points, approximately 31.25 with 160 fps
141# to be considered a peak. You can reduce this to 3 instead of 5 for 120 fps
142# Should be an odd number
143minimum_consecutive_points = 5
144
145index_above_threshold_clean = index_above_threshold.copy()
146# Erode by two points on either side
147# Remove two rising edges and two falling edges
148
149for i in range(1, minimum_consecutive_points, 2):
150 edges = np.diff(index_above_threshold_clean, prepend=False)
151 index_above_threshold_clean[edges] = False
152 edges = np.diff(index_above_threshold_clean, append=False)
153 index_above_threshold_clean[edges] = False
154
155
156for i in range(1, minimum_consecutive_points, 2):
157 edges = np.diff(index_above_threshold_clean, append=False)
158 index_above_threshold_clean[edges] = True
159 edges = np.diff(index_above_threshold_clean, prepend=False)
160 index_above_threshold_clean[edges] = True
161
162fig, ax = plt.subplots()
163ax.plot(time, speed * 1E3,
164 label="Zebrafish Speed (mm/s)")
165ax.plot([time[0], time[-1]], [speed_threshold * 1E3, speed_threshold * 1E3], '--r',
166 label=f"Threshold: {speed_threshold * 1E3:.1f} mm/s")
167ax.plot(
168 time[index_above_threshold],
169 speed[index_above_threshold] * 1E3, '.',
170 label="Speed Above Threshold (raw)",
171)
172ax.plot(
173 # time[index_above_threshold_clean],
174 time,
175 speed.where(index_above_threshold_clean) * 1E3, '-o',
176 label="Speed Above Threshold (filtered)",
177 markerfacecolor=(0, 0, 0, 0), markeredgecolor='m',
178 color="m")
179ax.set_ylabel("Speed (mm/s)")
180ax.set_xlabel("Time (s)")
181ax.set_title(f"Speed for zebrafish in well {well_name}")
182ax.legend()
183ax.grid()
184# %% Count the number of events:
185rising_edges = (
186 np.diff(index_above_threshold_clean, prepend=False) &
187 index_above_threshold
188)
189falling_edges = (
190 np.diff(index_above_threshold_clean, prepend=False) &
191 (~index_above_threshold)
192)
193
194time_event_start = rising_edges.index[rising_edges == True].to_numpy() # noqa
195time_event_end = falling_edges.index[falling_edges == True].to_numpy() # noqa
196number_of_events = len(time_event_end)
197event_duration = time_event_end - time_event_start
198average_duration = event_duration.mean()
199print(f"There were {number_of_events} events")
200print(f"The average duration of the events was {average_duration * 1E3:.0f} milliseconds.")
Zebrafish Thigmotaxis Assay Analysis¶
1"""
2%% Sample Thigmotaxis Assay Analysis
3By Ramona Optics Inc. Copyright 2022-2025
4
5This example script gives one method for using MCAM™ tracking data to analyze
6a Thigmotaxis assay using a 24-well plate
7
8According to one publication (Schnorr S., et. al, 2011), the authors
9validate a Thigmotaxis assay using a 24 well plate (16.2mm diameter wells)
10with both light/dark conditions and stimulant/depressant chemical
11experimental conditions. The authors comments (Materials and Methods
12Section 2.3, pg. 368) that well plate selection is determined by assuring
13the “swimming arena must be sufficiently large to allow distinction
14between inner and outer zones” and to do this both inner and outer
15zones must be “at least equivalent or larger than the body length of
16the larvae (approx 4mm for larvae aged 5dpf)”. It is also noted that
17a 6 well or 12 well format could work for this assay however using a
1824-well plate the area of inner and outer zones are equal “thus ruling
19out biases in the analysis of zone preference related to differences in
20zone size.” Finally they comment that both 96- and 48-well plate formats
21are likely too small to fit these requirements.
22
23Reference: Measuring thigmotaxis in larval zebrafish Schnorr et al. 2011
2410.1016/j.bbr.2011.12.016
25https://www.sciencedirect.com/science/article/abs/pii/S0166432811008758?via%3Dihub
26
27This assay has been formulated with the 24-well plate in mind. This script,
28specifically, determines the radius of the inner zone of a single well for
29each well of the well plate and checks if x, y locations of each fish are
30within this outside of this zone.
31
32Notes:
33 For this analysis we have chosen not to interpolate to fill gaps of non-
34 confident values. Only confident values are considered.
35
36Assay Output (.csv):
37Fraction confident frames
38Fraction of time spent in the outer zone for each well
39"""
40
41import numpy as np
42import pandas as pd
43
44# %% definitions:
45# define the filepath location of the previously output tracking data
46tracking_data_filename = 'path/tracking_data.csv'
47# define an output path for the data .csv file,
48# this should include a filename with '.csv' suffix
49output_filepath = 'path/thigmotaxis_data.csv'
50
51# the outer zone width, change this input parameter given in millimeters
52outer_zone_width_in_mm = 4
53# well plate single-well diameter, this may need to be changed depending on
54# the well plate in use, again given in millimeters
55well_diameter_in_mm = 16.2
56well_radius_in_mm = well_diameter_in_mm / 2
57inner_zone_radius_in_mm = well_radius_in_mm - outer_zone_width_in_mm
58inner_zone_radius_in_m = inner_zone_radius_in_mm / 1E3
59
60# set the confidence threshold used to filter out badly tracked keypoints
61# anecdotally 0.1 yields a very similar result to 0.95 while retaining
62# significantly more frames, we recommend using 0.1
63confidence_threshold = 0.1
64# select the keypoint to be tracked, generally 'center'
65tracked_keypoint = 'center'
66
67# %% load the previously exported raw tracking data
68tracking_data_df = pd.read_csv(
69 tracking_data_filename,
70 comment='#',
71 header=[0, 1, 2, 3],
72 index_col=0,
73)
74N_frames = len(tracking_data_df)
75
76# construct a list of well letter and number combinations that exist
77column_keys_array = np.array(list(tracking_data_df.keys()))
78well_letters = np.unique(column_keys_array[:, 0])
79well_numbers = np.unique(column_keys_array[:, 1])
80# ensure well number sorting occurs as if they are integers for proper ordering
81well_numbers = sorted(well_numbers, key=int)
82wellnames = []
83for well_letter in well_letters:
84 for well_number in well_numbers:
85 wellnames.append((well_letter, well_number))
86
87# construct an array to store output data
88output_data = np.zeros((2, len(wellnames)))
89
90# %% iterate through each well computing the time spent in the outer zone
91for i, well in enumerate(wellnames):
92 well_letter = well[0]
93 well_number = well[1]
94
95 x_key = (well_letter, well_number, tracked_keypoint, 'x')
96 y_key = (well_letter, well_number, tracked_keypoint, 'y')
97 likelihood_key = (well_letter, well_number, tracked_keypoint, 'likelihood')
98
99 # compute the fraction of confident frames
100 confident_frames = tracking_data_df[
101 tracking_data_df[likelihood_key] >= confidence_threshold]
102 N_confident_frames = len(confident_frames)
103 fraction_confident = N_confident_frames / N_frames
104
105 # compute the distance from origin at each timepoint
106 distance_from_origin = np.sqrt(confident_frames[x_key] ** 2 +
107 confident_frames[y_key] ** 2)
108 # count the frames the fish is in the outer zone
109 outer_zone_frames = len(confident_frames[
110 distance_from_origin > inner_zone_radius_in_m])
111 fraction_in_outer_zone = outer_zone_frames / N_frames
112
113 # store computed data to output data array
114 output_data[0, i] = fraction_confident
115 output_data[1, i] = fraction_in_outer_zone
116
117
118# %% construct an output dataframe with the data we have generated
119column_keys = pd.MultiIndex.from_tuples(wellnames)
120index = ['fraction_confident_frames', 'fraction_in_outer_zone']
121thigmotaxis_data_df = pd.DataFrame(
122 output_data,
123 index=index,
124 columns=column_keys
125)
126
127# %% save the output data
128thigmotaxis_data_df.to_csv(output_filepath, index=True)
Extracting Stimulus Metadata¶
1"""
2# %% Extract stimulus metadata from an MCAM dataset
3# By Ramona Optics Inc. Copyright 2023-2025
4
5Modify the metadata and output paths.
6
7This script assumes that stimulus information is present in the input metadata file.
8
9"""
10
11from pathlib import Path
12
13from owl import mcam_data
14from owl.analysis.tracking_data_analysis import export_csv, make_stimulus_metadata_dataframe
15
16metadata_path = '/path/to/metadata.nc'
17
18output_path = '/path/to/output_folder'
19
20metadata = mcam_data.load(metadata_path)
21output_path = Path(output_path)
22
23stimulus_metadata_df, header = make_stimulus_metadata_dataframe(
24 metadata,
25 row_index='time'
26)
27output_filepath = output_path / 'stimulus_metadata.csv'
28export_csv(
29 stimulus_metadata_df,
30 output_filepath,
31 header=header,
32 index=True
33)
34
35print(f"Stimulus metadata saved to {output_filepath}")
For more information, please see the section titled “MCAM Data Analysis”
in the MCAM User Manual.
Pulse power calibration¶
It may be desirable for a variety of experiments to calibrate the pulse power from the flash stimulus. Here we provide an example of how to calibrate the approximate optical power delivered to the animals during a stimulus event delivered by the fluorescence LEDs.
Note that absolute measured intensity will vary based on the distance between the animals and the light source, as well as the exact wavelength used for illumination.
The average power will also be a function of the pulse duration. The LEDs can take up to 200 ms to reach their peak power, and as they heat up the power will decrease.
As of Version 0.19.135, 1% fluorescence brightness is equivalent 1,000 lux as displayed on the interface.
In the following setup, we used a Thorlabs PM100A power meter with a S120C sensor to measure the power at the z-stage location where a wellplate would be in behavior experiments. The sample setup is shown in the image below.

We then increased the intensity of the light source in 10% increments and recorded the measured peak power for 1s pulses.
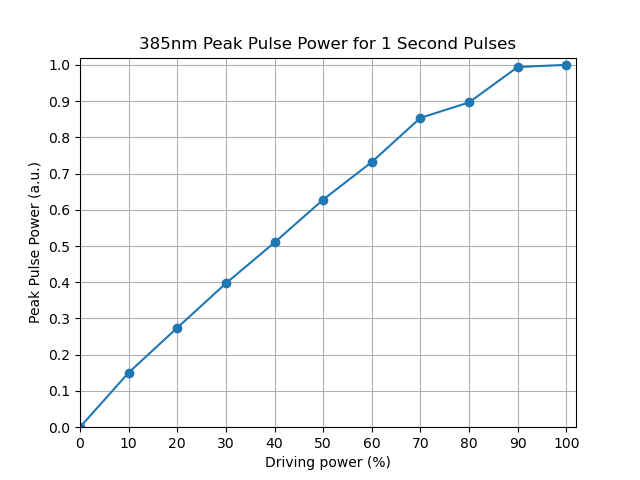
As one can see, the power increases linearly with the intensity saturating roughly at 90%.
Note that while the S120C’s responsivity is not specifically calibrated for 385 nm, the weak response of the photodiode can still be used to estimate the linearity of the flash stimulus.